
September 11, 2018
In life, timing is everything.
Your body’s internal clock – the circadian rhythm – regulates an enormous variety of processes: when you sleep and wake, when you’re hungry, when you’re most productive. Given its palpable effect on so much of our lives, it’s not surprising that it has an enormous impact on our health as well. Researchers have linked circadian health to the risk of diabetes, cardiovascular disease and neurodegeneration. It’s also known that the timing of meals and medicines can influence how they’re metabolized.
The ability to measure one’s internal clock is vital to improving health and personalizing medicine. It could be used to predict who is at risk for disease and track recovery from injuries. It can also be used to time the delivery of chemotherapy and blood pressure and other drugs so that they have the optimum effect at lower doses, minimizing the risk of side effects.
However, reading one’s internal clock precisely enough remains a major challenge in sleep and circadian health. The current approach requires taking hourly samples of blood melatonin – the hormone that controls sleep – during day and night, which is expensive and extremely burdensome for the patient. This makes it impossible to incorporate into routine clinical evaluations.
My colleagues and I wanted to obtain precise measurements of internal time without the need for burdensome serial sampling. I’m a computational biologist with a passion for using mathematical and computational algorithms to make sense of complex data. My collaborators, Phyllis Zee and Ravi Allada, are world-renowned experts in sleep medicine and circadian biology. Working together, we designed a simple blood test to read a person’s internal clock.
The circadian rhythm is present in every single cell of your body, guided by the central clock that resides in the suprachiasmatic nucleus region of the brain. Like the secondary clocks in an old factory, these so-called “peripheral” clocks are synchronized to the master clock in your brain, but also tick forward on their own – even in petri dishes!
Your cells keep time through a network of core clock genes that interact in a feedback loop: When one gene turns on, its activity causes another molecule to turn it back down, and this competition results in an ebb and flow of gene activation within a 24-hour cycle. These genes in turn regulate the activity of other genes, which also oscillate over the course of the day. This mechanism of periodic gene activation orchestrates biological processes across cells and tissues, allowing them to take place in synchrony at specific times of day.
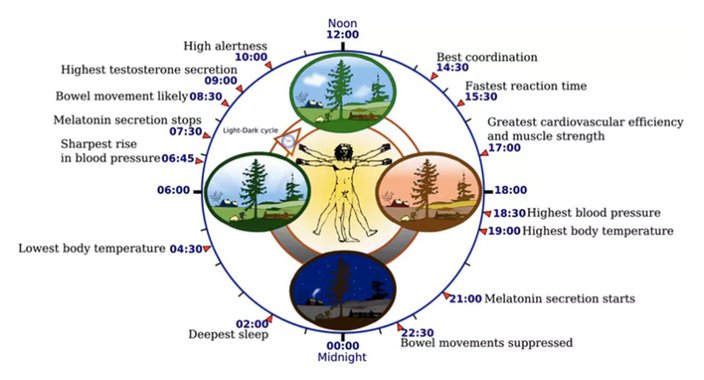 Yassine Mrabet/CC BY-SA
Yassine Mrabet/CC BY-SAThe circadian rhythm orchestrates many biological processes, including digestion, immune function and blood pressure, all of which rise and fall at specific times of day. Misregulation of the circadian rhythm can have adverse effects on metabolism, cognitive function and cardiovascular health.
The discovery of the core clock genes is so fundamental to our understanding of how biological functions are orchestrated that it was recognized by the Nobel Committee last year. Jeffrey C. Hall, Michael Rosbash and Michael W. Young together won the 2017 Nobel Prize in Physiology or Medicine “for their discoveries of molecular mechanisms controlling the circadian rhythm.” Other researchers have noted that as many as 40 percent of all other genes respond to the circadian rhythm, changing their activity over the course of the day as well.
This gave us an idea: Perhaps we could use the activity levels of a set of genes in the blood to deduce a person’s internal time – the time your body thinks it is, regardless of what the clock on the wall says. Many of us have had the experience of feeling “out of sync” with our environments – of feeling like it’s 5:00 a.m. even though our alarm insists it’s already 7:00. That can be a result of our activities being out of sync with our internal clock – the clock on the wall isn’t always a good indication of what time it is for you personally. Knowing what a profound impact one’s internal clock can have on biology and health, we were inspired to try to gauge gene activity to measure the precise internal time in an individual’s body. We developed TimeSignature: a sophisticated computational algorithm that could measure a person’s internal clock from gene expression using two simple blood draws.
To achieve our goals, TimeSignature had to be easy (measuring a minimal number of genes in just a couple blood draws), highly accurate and – most importantly – robust. That is, it should provide just as accurate a measure of your intrinsic physiological time regardless of whether you’d gotten a good night’s sleep, recently returned from an overseas vacation or were up all night with a new baby. And it needed to work not just in our labs but in labs across the country and around the world.
To develop the gene signature biomarker, we collected tens of thousands of measurements every two hours from a group of healthy adult volunteers. These measurements indicated how active each gene was in the blood of each person during the course of the day. We also used published data from three other studies that had collected similar measurements. We then developed a new machine learning algorithm, called TimeSignature, that could computationally search through this data to pull out a small set of biomarkers that would reveal the time of day. A set of 41 genes was identified as being the best markers.
Surprisingly, not all the TimeSignature genes are part of the known “core clock” circuit – many of them are genes for other biological functions, such as your immune system, that are driven by the clock to fluctuate over the day. This underscores how important circadian control is – its effect on other biological processes is so strong that we can use those processes to monitor the clock!
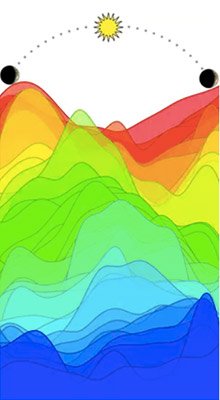 Source/ Rosemary Braun
Source/ Rosemary BraunMany genes peak in activity at different times of day. This set of 41 genes, each shown as a different color, shows a robust wave of circadian expression. By monitoring the level of each gene relative to the others, the TimeSignature algorithm learns to ‘read’ your body’s internal clock.
We then tested our TimeSignature algorithm by applying it to the remaining data, and demonstrated that it was highly accurate: We were able to deduce a person’s internal time to within 1.5 hours. We also demonstrated our algorithm works on data collected in different labs around the world, suggesting it could be easily adopted. We were also able to demonstrate that our TimeSignature test could detect a person’s intrinsic circadian rhythm with high accuracy, even if they were sleep-deprived or jet-lagged.
By making circadian rhythms easy to measure, TimeSignature opens up a wide range of possibilities for integrating time into personalized medicine. Although the importance of circadian rhythms to health has been noted, we have really only scratched the surface when it comes to understanding how they work. With TimeSignature, researchers can now easily include highly accurate measures of internal time in their studies, incorporating this vital measurement using just two simple blood draws. TimeSignature enables scientists to investigate how the physiological clock impacts the risk of various diseases, the efficacy of new drugs, the best times to study or exercise and more.
Of course, there’s still a lot of work to be done. While we know that circadian misalignment is a risk factor for disease, we don’t yet know how much misalignment is bad for you. TimeSignature enables further research to quantify the precise relationships between circadian rhythms and disease. By comparing the TimeSignatures of people with and without disease, we can investigate how a disrupted clock correlates with disease and predict who is at risk.
Down the road, we envision that TimeSignature will make its way into your doctor’s office, where your circadian health could be monitored just as quickly, easily and accurately as a cholesterol test. Many drugs, for example, have optimal times for dosing, but the best time for you to take your blood pressure medicine or chemotherapy may differ from somebody else.
Previously there was no clinically feasible way to measure this, but TimeSignature makes it possible for your doctor to do a simple blood test, analyze the activity of 41 genes and recommend the time that would give you the most effective benefits. We also know that circadian misalignment – when your body’s clock is out of sync with the external time – is a treatable risk factor for cognitive decline; with TimeSignature, we could predict who is at risk, and potentially intervene to align their clocks.![]()
Rosemary Braun, Assistant Professor of Biostatistics, Preventive Medicine and Engineering Sciences and Applied Mathematics, Northwestern University
This article is republished from The Conversation under a Creative Commons license. Read the original article.